Abstract
Response heterogeneity to targeted therapies is a major barrier to achieving optimal outcomes in a broad range of myeloproliferative neoplasms (MPN) with druggable oncogenes. This remains the case in chronic myeloid leukemia (CML) where pharmacologic targeting of the causative BCR-ABL1 kinase with tyrosine kinase inhibitors (TKI) elicits highly heterogeneous responses. The reasons why some patients enjoy deep molecular responses while others progress to blast crisis (BC) remain unknown. CML is an ideal model to study primary drug resistance since the disease is defined and driven by BCR-ABL1, treated with potent ABL1 inhibitors, and molecular response monitoring is standard.
To characterize the cellular and molecular features of primary TKI resistance, we performed single-cell multi-omics (scRNA-seq and CyTOF) and mutational analysis of pretreatment chronic phase (CP) bone marrow (BM) mononuclear cells (MNC) from three prognostic Groups: patients with major molecular responses (MMR) to first-line imatinib (IM) (Group A, n=8), sub-optimal responses to IM but favorable responses to 2nd/3rd-line TKIs (Group B, n=9); and pan-TKI resistance with eventual BC progression (Group C, n=6).
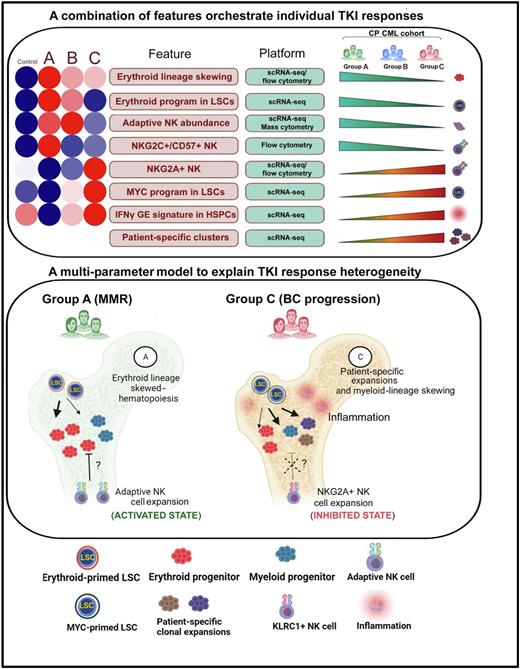
We generated a single-cell atlas of 163,146 MNCs, which allowed us to identify 8 statistically significant features in pretreatment BM correlating with either IM sensitivity (MMR) or increasing resistance (Figure, top panel). Next, using an unbiased machine-learning approach, we identified leukemia stem cell (LSC) and NK cell gene expression (GE) profiles predicting IM response with >80% accuracy.
To better understand the functional implications of these GE signatures, we further characterized the LSC and NK cell populations computationally and experimentally. To determine the contribution of LSC fate decisions, we employed the SCENIC algorithm to identify differential LSC regulon activity between normal BM and Groups A-C. Compared to normal BM, we found an increase in the activity of a canonical erythroid-specifying regulon (TAL1/KLF1/GATA1) in Group A LSCs, which diminished with increasing TKI resistance. Both in vitroand in vivo readouts supported the functional activity of the erythroid regulon, observable as a marked (6.3-fold in Group A vs 1.09-fold in Group C) erythroid over myeloid bias in colony forming assays, and erythroid progenitor (ERP) expansion in Group A vs C BM (p<0.05) respectively. Importantly, ERPs demonstrated exquisite TKI sensitivity compared to myeloid progenitors (p<0.001). The LSC features were lost with progressive resistance, and in patients who transformed, MYC- and IRF1-driven inflammatory regulons became evident.
We next performed de novo clustering of 8,617 NK cells, and observed a 56-fold expansion (p<0.01) of a normally rare subset of hyperfunctional adaptive NK cells in Group A patients that diminished with progressive resistance. We also characterized signaling interactions between stem and progenitor cell (SPC) ligands and NK cell receptors. We found that Group A NK-SPC interactions were notable for enrichment and depletion of multiple activating and inhibitory interactions respectively. In contrast, comparing Group C vs A NK-SPC interactions, we found a striking upregulation of a key inhibitory interaction between NKG2A and HLA-E.
We next assessed the interplay between genetic mutations [copy number and single nucleotide variants (CNV/SNV)] and patient-specific cluster expansions. We found that while CNV-driven clonal expansion within CML hierarchies is a marker for IM resistance, the commonest recurrent mutations in CML, including ASXL1, TET2, and SETD1B, were not associated with an increased risk of BC.
Finally, and as a step toward developing clinical biomarkers, we validated our scRNA-seq findings with a custom antibody set by CyTOF and conventional flow cytometry, and found that they faithfully replicated the cell abundance gradients and GE patterns uncovered by our single cell atlas.
In summary, we describe a comprehensive multi-modal analysis of the genetic, cellular and molecular features of primary TKI resistance in CML, and provide novel insights into the complex interactions between LSCs, immune cells, inflammation, and genetic mutations contributing to response heterogeneity (Figure, bottom panel). We envisage that our findings may be relevant to understanding primary resistance to targeted therapies in other MPNs.
Disclosures
Chuah:Pfizer: Other: Travel, Research Funding; Steward Cross: Korea Otsuka International Asia Arab: Honoraria; Otsuka [Philippines] Pharmaceutical: Honoraria; Bristol Myers Squibb: Honoraria, Research Funding; Korea Otsuka Pharmaceutical: Honoraria; Novartis: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.